This part presents the alternative splicing caused peptide identification result.
Figure 1. Junction type.

Table 1. Get Full Table Alternative splicing caused peptide identification.
| peptide | ID | isUnique | junType | Query | Qvalue | rt |
|---|---|---|---|---|---|---|
| EEAVQQMADALQYLQK |
JUC31972 JUC5413 JUC5415 JUC9418 |
TRUE |
connect a known exon and a non-exon region connect a known exon and a region overlap with known exon novel alternative splicing junction novel alternative splicing junction |
317494 | 0.0021 | 7826.57 |
| EEPFVADEYIER | JUC34312 | TRUE | connect a known exon and a non-exon region |
238162 238163 |
0 0 |
6455.68 6458.86 |
| PLADAHLTK | JUC70253 | TRUE | connect a known exon and a non-exon region |
81195 81196 |
4e-04 4e-04 |
3647.14 3645.19 |
| SLLCDPNPDDPLVPEIAR | JUC73502 | TRUE | connect a known exon and a non-exon region |
345704 345707 345705 345706 |
0 0 0 4e-04 |
7286.48 7341.67 7339.31 7283.6 |
| LGEVEDSTMPIR | JUC472 | TRUE | connect a known exon and a region overlap with known exon |
200684 200686 |
0 0 |
4651.15 4648.49 |
| LMLGGGGYTIR | JUC27255 | TRUE | connect a known exon and a region overlap with known exon |
134695 134696 |
4e-04 4e-04 |
5190.02 5188.07 |
| WLGTPIEEMR |
JUC30008 JUC30037 JUC30064 |
TRUE |
connect a known exon and a region overlap with known exon connect a known exon and a region overlap with known exon connect a known exon and a region overlap with known exon |
165472 165467 |
0 0 |
5846.33 5848.31 |
| FEESKEPVADEEEEDSDDDVEPITEFR | JUC5002 | TRUE | connect a known exon and a region overlap with known exon | 457145 | 0 | 6461.85 |
| AQEDALAQQAFEEAR | JUC37198 | TRUE | connect a known exon and a region overlap with known exon | 278481 | 0 | 6132.09 |
| ETWVEEDELFQVQAPDEDSTTNITK | JUC63829 | TRUE | connect a known exon and a region overlap with known exon |
444209 444210 |
0 5e-04 |
7783.06 7778.79 |
| DDDEPGPWTK | JUC12901 | TRUE | connect a known exon and a region overlap with known exon |
141557 141558 |
0.0014 0.0049 |
4397.92 4399.96 |
| RDDDEPGPWTK | JUC12901 | TRUE | connect a known exon and a region overlap with known exon |
191139 191138 |
8e-04 0.0011 |
3685.56 3683.14 |
| LANQDEGPEDEEDEQNSPVAPTAQPK | JUC66211 | TRUE | connect a known exon and a region overlap with known exon |
436975 436976 |
0 0 |
4380.22 4377.28 |
| SSQEVVDFIQSK | JUC13914 | TRUE | connect a known exon and a region overlap with known exon | 206468 | 0.0043 | 5284.34 |
| AVDAVIAELKK | JUC15200 | TRUE | connect a known exon and a region overlap with known exon | 140785 | 0.004 | 8995.17 |
| CAPVMVELEGETDPLLIAMK | JUC43557 | TRUE | connect a known exon and a region overlap with known exon |
376510 376511 |
5e-04 0.0011 |
9373.78 9376.7 |
| SEVADFEPER | JUC45318 | TRUE | connect a known exon and a region overlap with known exon |
147946 147945 |
0.0014 0.0014 |
4682.19 4684.31 |
| AALLGNEDTAVEEPVPEVVPVQVETAK | JUC46073 | TRUE | connect a known exon and a region overlap with known exon |
436737 436736 |
0.0011 0.0028 |
7386.97 7389.47 |
| EEEMQVDQEEPHVEEQQQQTPAENK | JUC20303 | TRUE | connect a known exon and a region overlap with known exon |
448780 448781 |
0 0 |
4507.72 4510.03 |
| PLILQSLGPK | JUC21479 | TRUE | connect a known exon and a region overlap with known exon |
112011 112013 112010 112012 |
0 0 0 0 |
5876.55 5869.87 6166.88 6169.46 |
| TLLEALDCILPPTRPTDKPLR | JUC74675 | TRUE | connect a known exon and a region overlap with known exon | 400532 | 0.0054 | 7713.56 |
| KVVGCSCVVVKK | JUC48464 | TRUE | connect a known exon and a region overlap with known exon | 205488 | 0 | 3015.32 |
| LLNLCEAEDSALTK | JUC49755 | TRUE | connect a known exon and a region overlap with known exon | 256809 | 0 | 5935.45 |
| RDPTVHDDVLELEMDELNR | JUC77120 | TRUE | connect a known exon and a region overlap with known exon |
387055 387056 |
0 0 |
6877.83 6880.34 |
| ENEQGEVDMESHR | JUC52293 | TRUE | connect a known exon and a region overlap with known exon |
256470 252977 252978 252976 252975 256472 256471 256469 |
0 0 0 5e-04 5e-04 5e-04 0.0011 0.0014 |
3012.22 3227.63 3416.9 3418.74 3229.96 3435.65 3437.56 3001.02 |
| GGGESLQEASPR | JUC79025 | TRUE | connect a known exon and a region overlap with known exon |
150878 150886 |
0 0.0014 |
3234.08 3231.57 |
| GTQEAEEEGSEATPEAGPEGQETAEITDFQR | JUC13487 | TRUE | connect two non-exon region |
462042 462041 |
0 0 |
6102.14 6126.49 |
| EDMAALEKDYEEVGVDSVE | JUC32417 | TRUE | connect two regions overlaped with known exons |
362933 362935 362934 362931 362936 |
0 5e-04 0.0011 0.0011 0.0011 |
7613.82 7617.37 7599.22 7602.64 7614.64 |
| ERGMEEGEFSEAR | JUC32417 | TRUE | connect two regions overlaped with known exons |
244873 244876 248873 248866 248868 248870 248867 248869 244874 244875 244877 |
0 0 0 0 0 0 0 0 0 4e-04 0.0018 |
4872.78 4875.36 4081.26 4227.4 4222.36 4079.26 4251.81 4951.69 4997.16 4999.68 5170.92 |
| ERGMEEGEFSEAREDMAALEK | JUC32417 | TRUE | connect two regions overlaped with known exons |
400042 400039 |
0.0011 0.0011 |
7316.75 7323.22 |
| FAHIDGDHLTLLNVY | JUC18888 | TRUE | connect two regions overlaped with known exons |
289374 289376 |
0.0028 0.0057 |
7023.77 7021.2 |
| MGEEEALQSQVTK | JUC52188 | TRUE | connect two regions overlaped with known exons | 227154 | 0 | 4623.04 |
| LSIRPNIQNPDPAVYQLR | JUC60611 | TRUE | gene fusion |
357299 357300 |
5e-04 5e-04 |
5793.48 5791.65 |
| VGNMSESELGR | JUC53574 | TRUE | novel alternative splicing junction |
147959 147962 |
0 0 |
3160.49 3162.51 |
| PILLQGHER | JUC53803 | TRUE | novel alternative splicing junction |
111003 111007 |
4e-04 0.0014 |
3076.73 3074.73 |
| DGYELSPTAAANFTR | JUC27308 | TRUE | novel alternative splicing junction |
264692 264691 264687 264690 264688 264689 |
0 0 0 0 0 0 |
5733.49 5731.26 5991.42 5952.89 5989.23 5955.91 |
| AAYAHELPK | JUC27845 | TRUE | novel alternative splicing junction |
91406 91405 |
0.0057 0.0065 |
2832.55 2830.51 |
| DGGNPFAEPSELDNPFQASAAAATAELLK | JUC28521 | TRUE | novel alternative splicing junction |
444836 444837 |
0 0.0011 |
9866.42 9869.3 |
| LQLVCHVSGFYPK | JUC28761 | TRUE | novel alternative splicing junction |
250275 250287 250285 250278 |
5e-04 0.0024 0.0011 0.0011 |
5414.51 5437.4 5440.28 5417.03 |
| LQEYGSIFTGAQDPGLQR | JUC62542 | FALSE | novel alternative splicing junction |
339092 339091 |
0 0 |
5991.1 5993.16 |
| HPWDDISYVLPEHM | JUC11718 | TRUE | novel alternative splicing junction |
291571 291570 |
0.0014 0.0057 |
7630.92 7632.82 |
| LLELDSFLKPTYK | JUC11761 | TRUE | novel alternative splicing junction | 254741 | 0 | 7239.13 |
| KNCQINSALTSFHTALELAVDQR | JUC65723 | TRUE | novel alternative splicing junction |
421788 421789 |
0.0038 0.0071 |
9229.89 9234.39 |
| GFWYDAEISR | JUC66024 | TRUE | novel alternative splicing junction |
169125 169124 |
0.0024 0.0046 |
5976.72 5979.34 |
| NDASEVVLAGER | JUC66024 | TRUE | novel alternative splicing junction |
174394 174395 |
0 0 |
4528.95 4531.41 |
| PYVEEGLHPQIIIR | JUC67353 | TRUE | novel alternative splicing junction |
276065 276066 276064 276063 |
0 5e-04 0.0011 0.0025 |
5325.79 5323.32 5201.71 5196.63 |
| EENPEGPPNANEDYR | JUC15000 | TRUE | novel alternative splicing junction |
289893 289888 |
0 0 |
3748.5 3750.5 |
| VKEENPEGPPNANEDYR | JUC15000 | TRUE | novel alternative splicing junction |
334950 334948 |
0 0.0011 |
3504.93 3502.9 |
| TAEGLLNVIGMDKPLTLPDFLAK | JUC69093 | TRUE | novel alternative splicing junction | 404652 | 5e-04 | 9790.5 |
| NLEALALDLMEPEQAVDLTYNEGSGSK | JUC70251 | TRUE | novel alternative splicing junction | 443171 | 0 | 10122.2 |
| SFENPVLQQHFR | JUC70251 | TRUE | novel alternative splicing junction |
239347 239343 |
0 0 |
5130 5132.54 |
| SCMVEESGTLESQLEATK | JUC52024 | TRUE | novel alternative splicing junction | 342225 | 0 | 6073.8 |
| TTYFEIFTAACNPSACR | JUC26445 | TRUE | novel alternative splicing junction |
343782 343783 |
4e-04 0.0043 |
7197.98 7195.35 |
Figure S1. Get High-res Image Peak annotation.

Figure S2. Get High-res Image Peak annotation.

Figure S3. Get High-res Image Peak annotation.
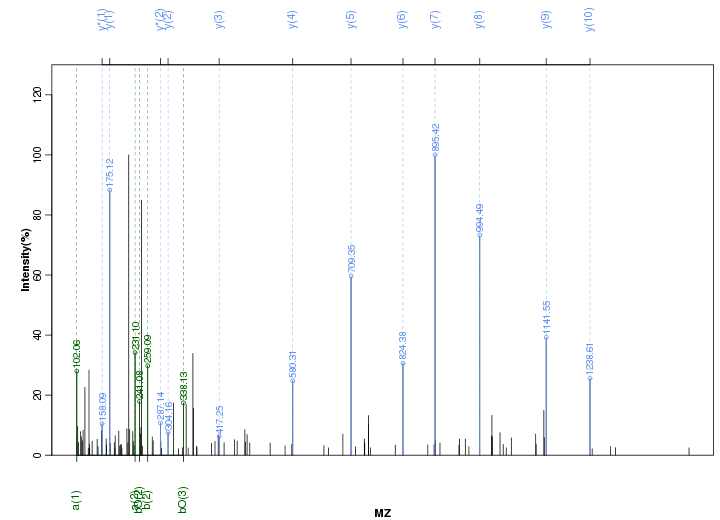
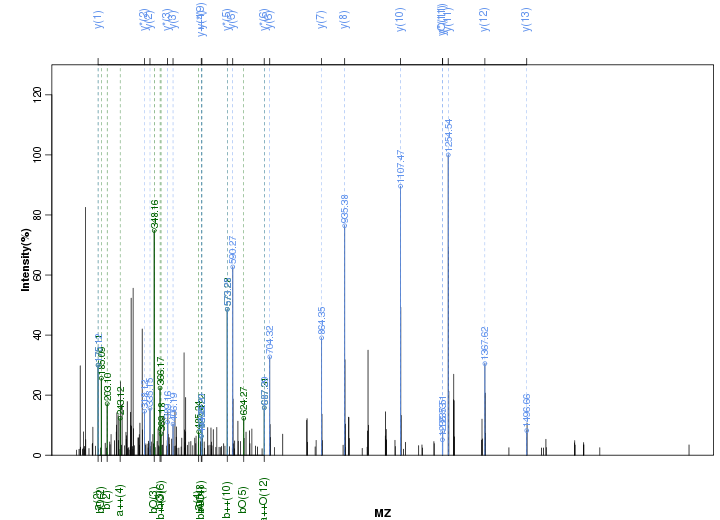
Figure S4. Get High-res Image Peak annotation.
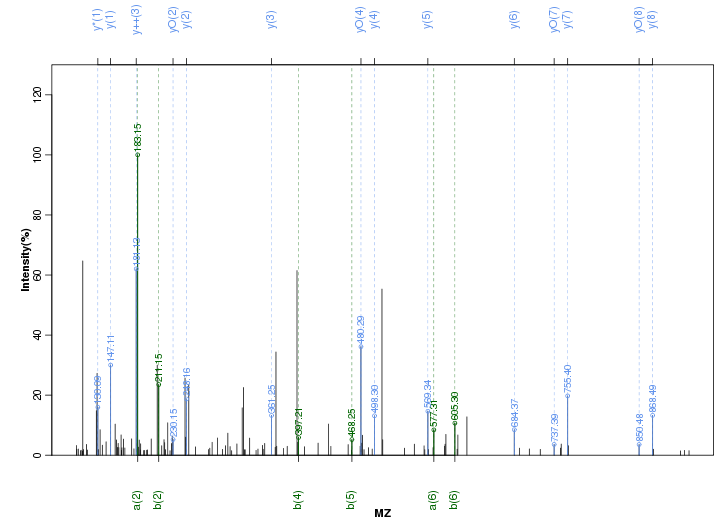
Figure S5. Get High-res Image Peak annotation.
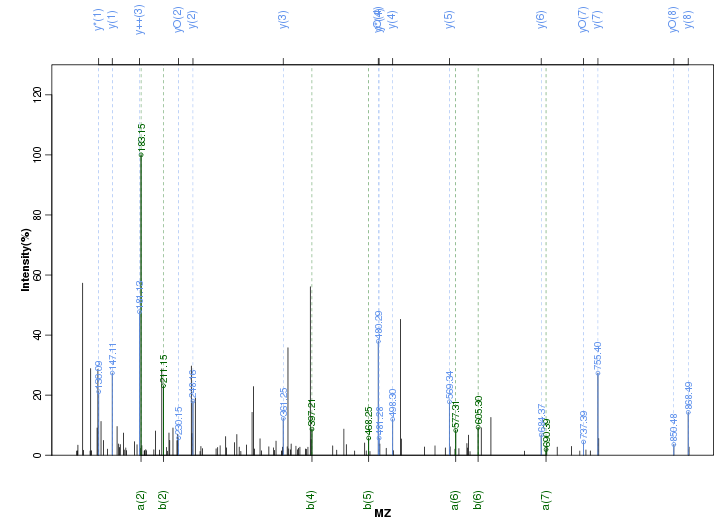
Figure S6. Get High-res Image Peak annotation.
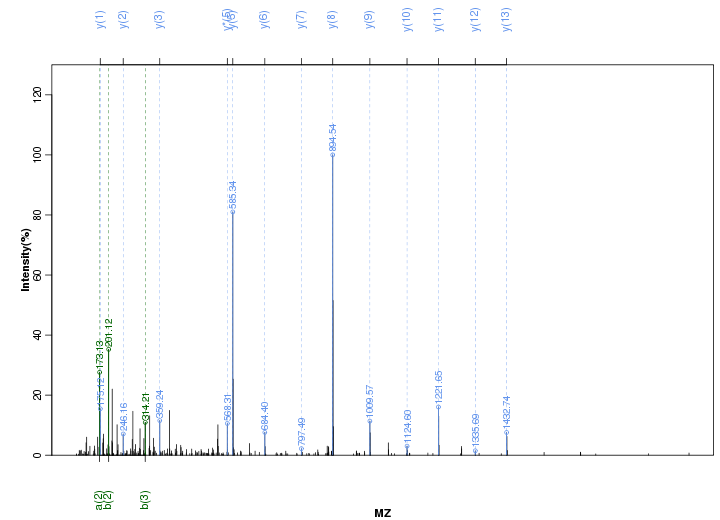
Figure S7. Get High-res Image Peak annotation.
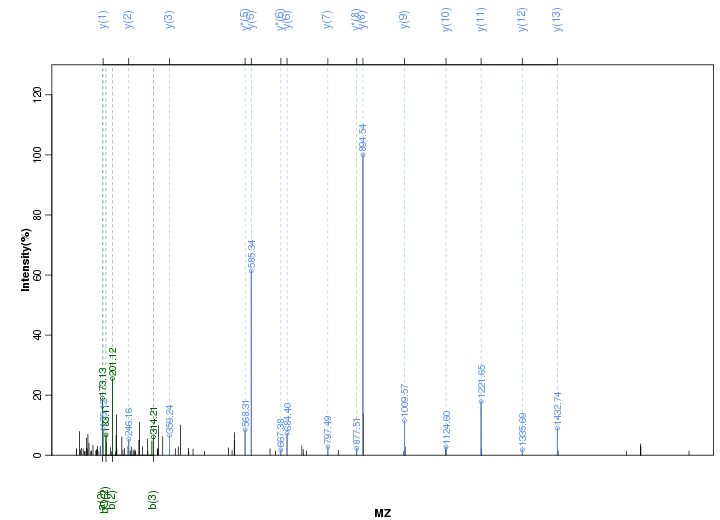
Figure S8. Get High-res Image Peak annotation.

Figure S9. Get High-res Image Peak annotation.
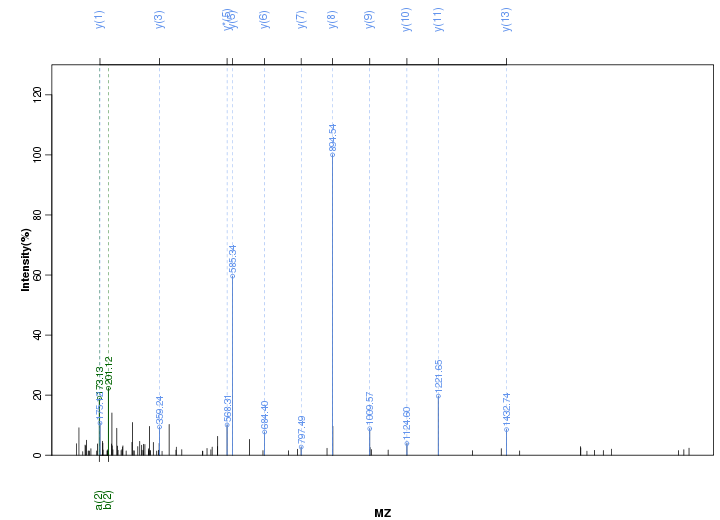
Figure S10. Get High-res Image Peak annotation.
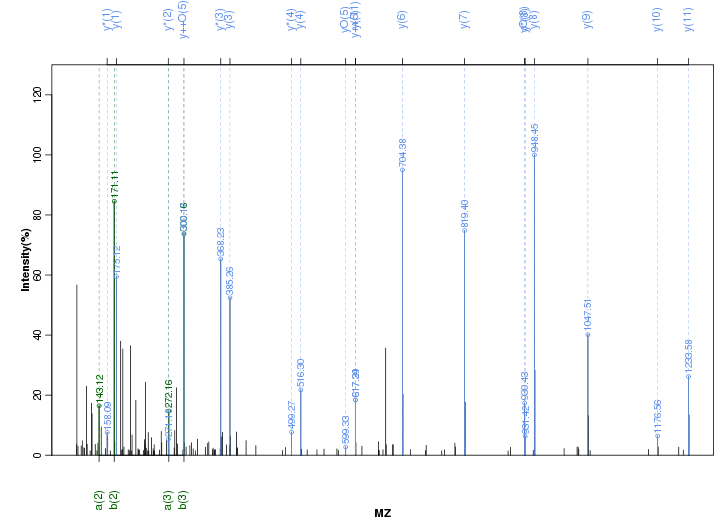
Figure S11. Get High-res Image Peak annotation.

Figure S12. Get High-res Image Peak annotation.
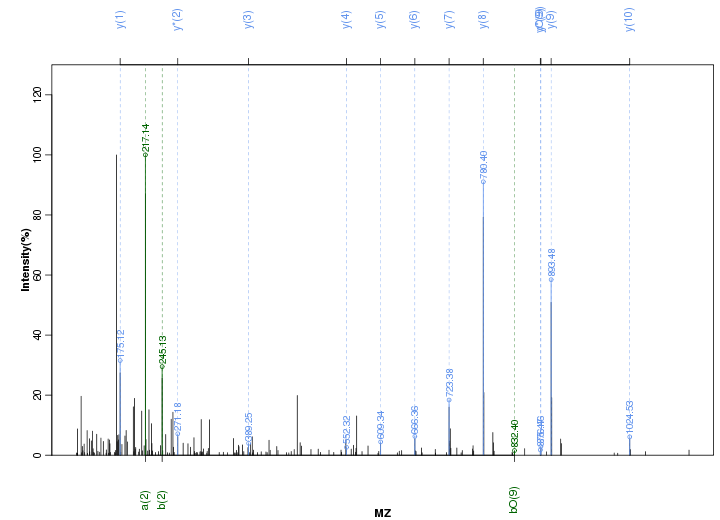
Figure S13. Get High-res Image Peak annotation.
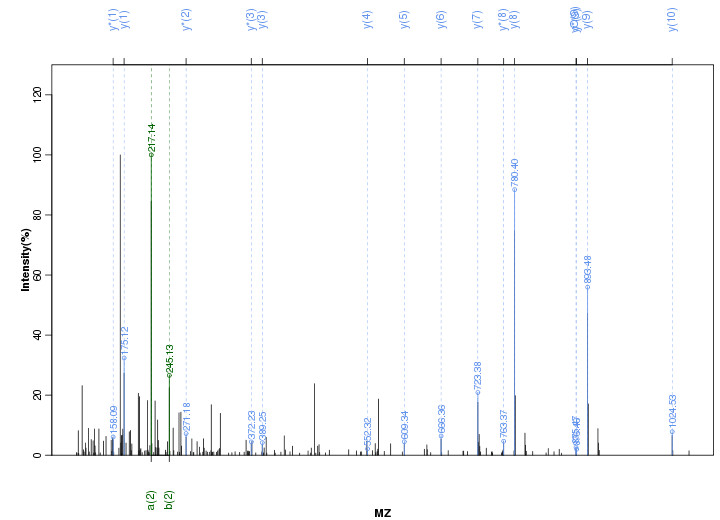
Figure S14. Get High-res Image Peak annotation.

Figure S15. Get High-res Image Peak annotation.
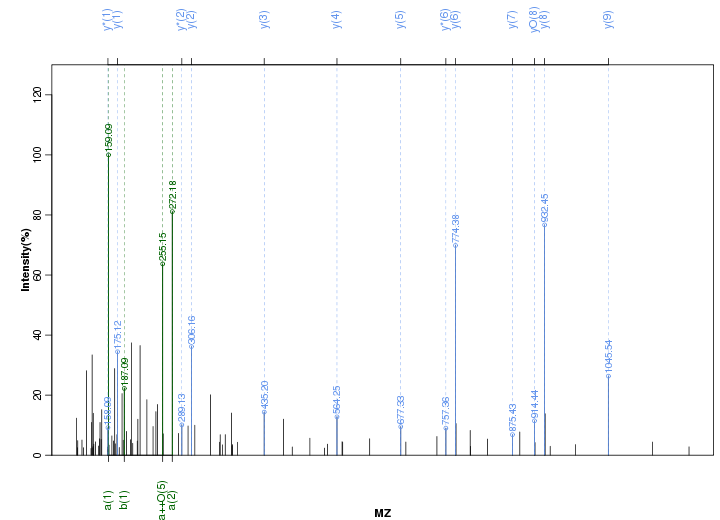
Figure S16. Get High-res Image Peak annotation.

Figure S17. Get High-res Image Peak annotation.
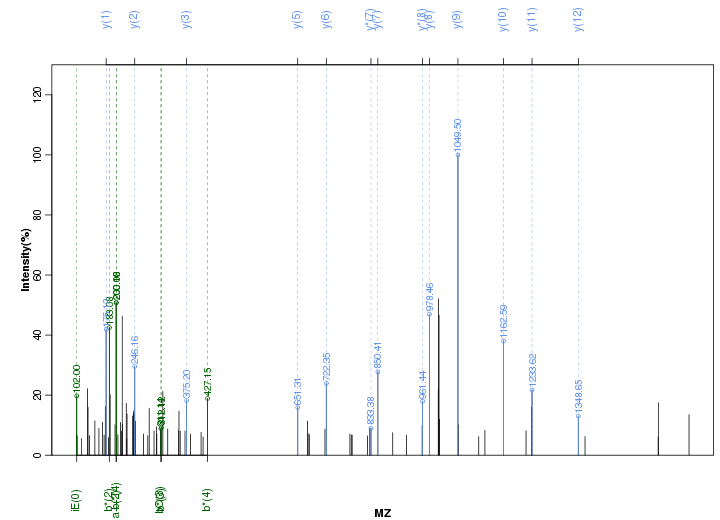
Figure S18. Get High-res Image Peak annotation.
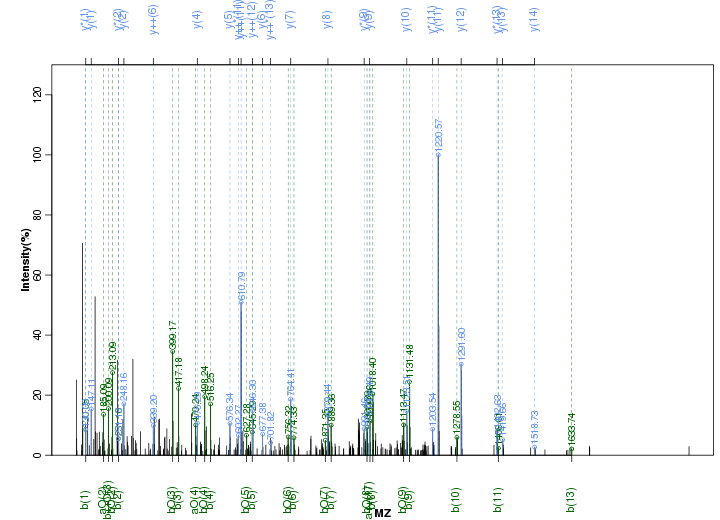
Figure S19. Get High-res Image Peak annotation.
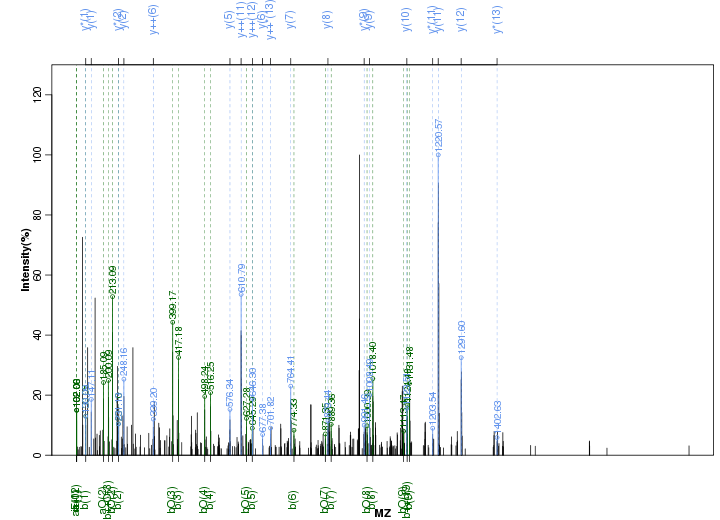
Figure S20. Get High-res Image Peak annotation.
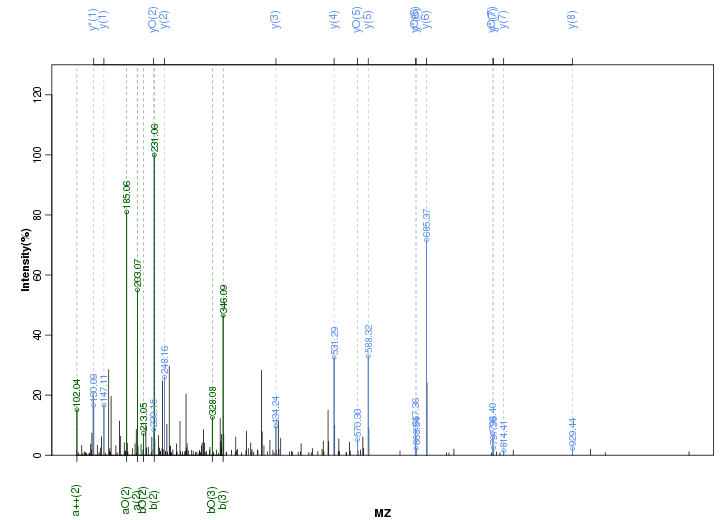
Figure S21. Get High-res Image Peak annotation.

Figure S22. Get High-res Image Peak annotation.
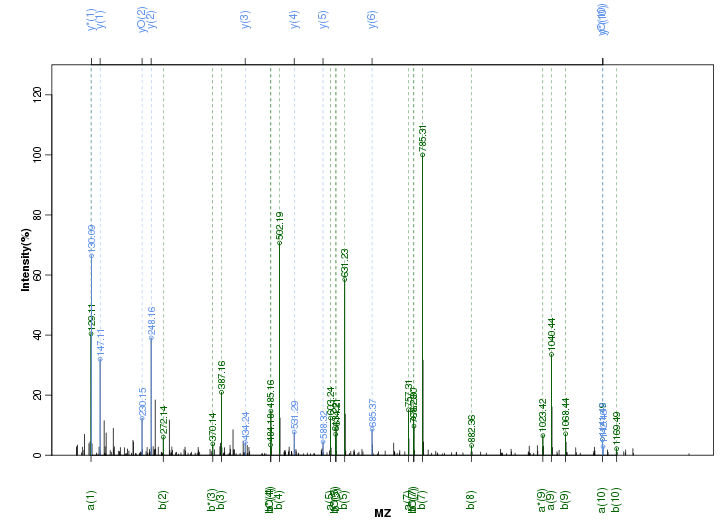
Figure S23. Get High-res Image Peak annotation.

Figure S24. Get High-res Image Peak annotation.
Figure S25. Get High-res Image Peak annotation.
Figure S26. Get High-res Image Peak annotation.
Figure S27. Get High-res Image Peak annotation.

Figure S28. Get High-res Image Peak annotation.
Figure S29. Get High-res Image Peak annotation.

Figure S30. Get High-res Image Peak annotation.

Figure S31. Get High-res Image Peak annotation.
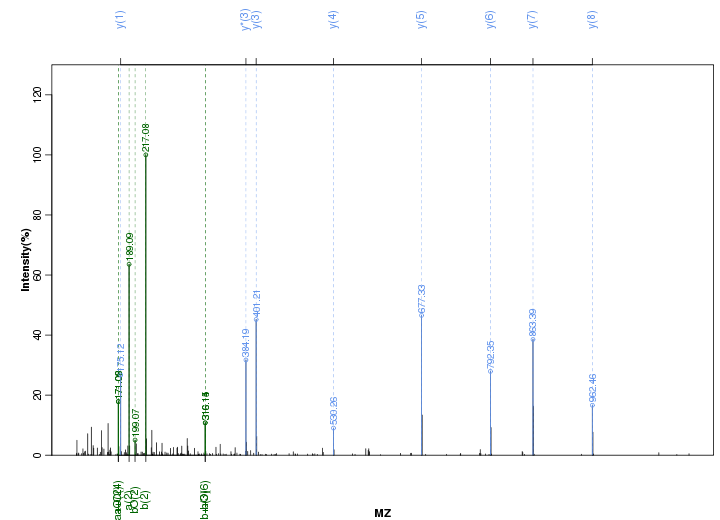
Figure S32. Get High-res Image Peak annotation.
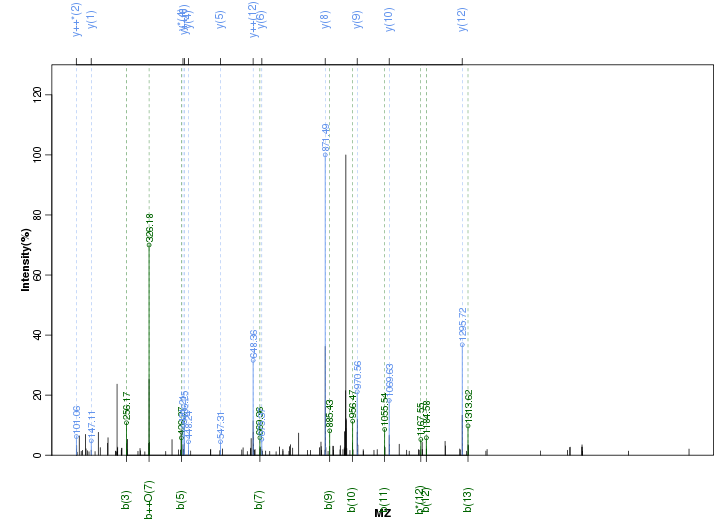
Figure S33. Get High-res Image Peak annotation.
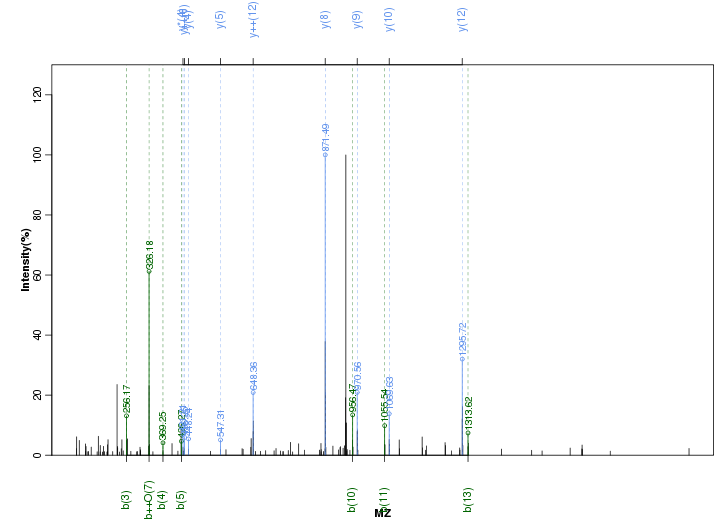
Figure S34. Get High-res Image Peak annotation.
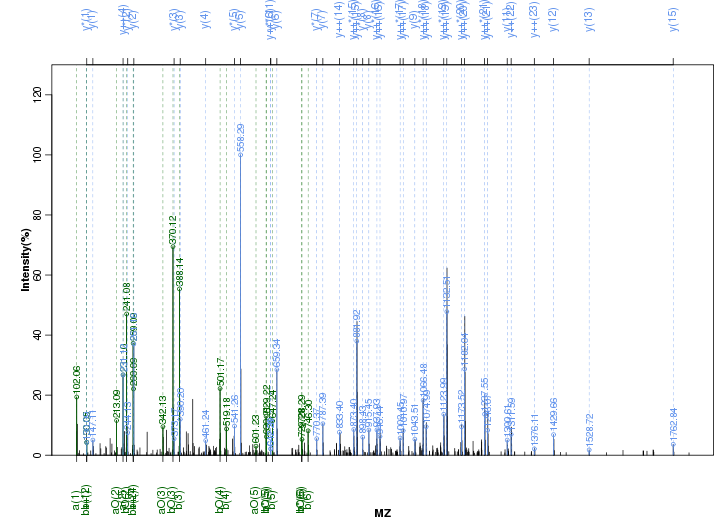
Figure S35. Get High-res Image Peak annotation.
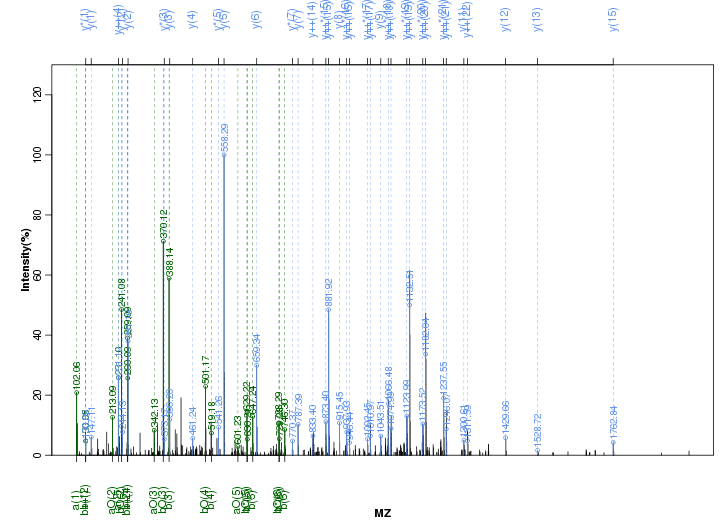
Figure S36. Get High-res Image Peak annotation.
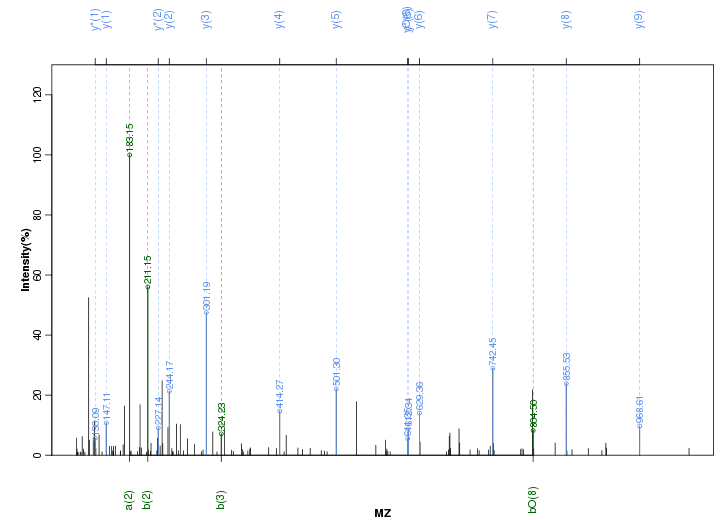
Figure S37. Get High-res Image Peak annotation.

Figure S38. Get High-res Image Peak annotation.

Figure S39. Get High-res Image Peak annotation.

Figure S40. Get High-res Image Peak annotation.
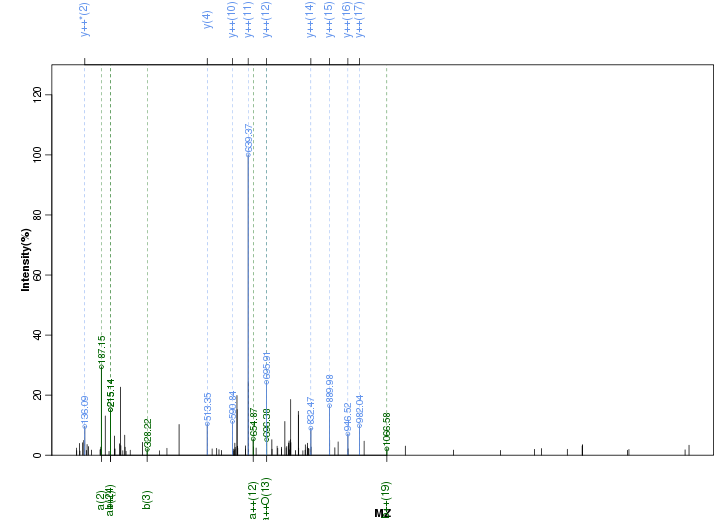
Figure S41. Get High-res Image Peak annotation.
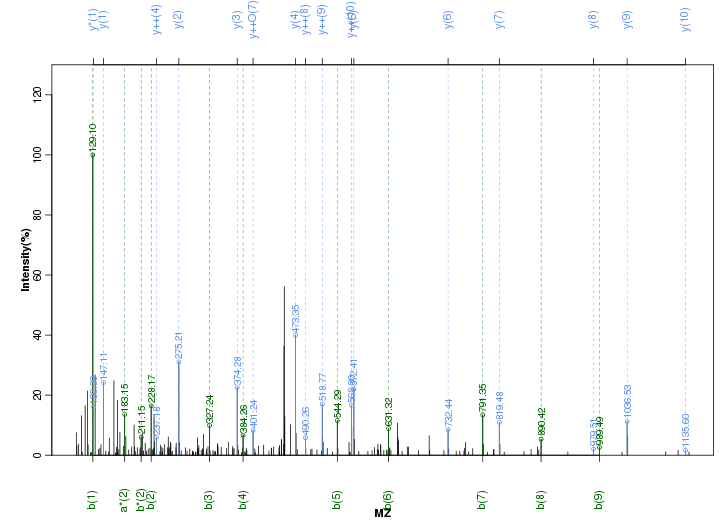
Figure S42. Get High-res Image Peak annotation.
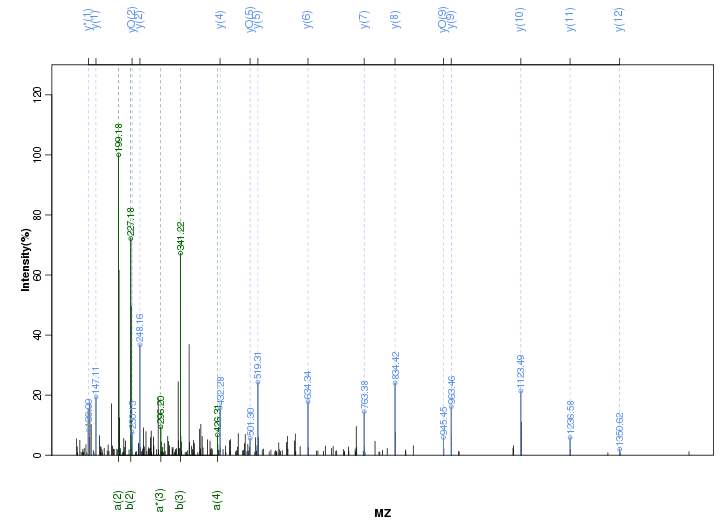
Figure S43. Get High-res Image Peak annotation.

Figure S44. Get High-res Image Peak annotation.
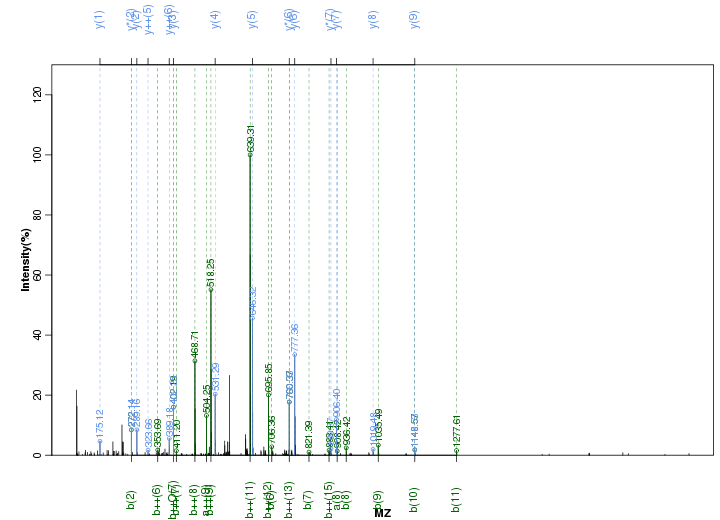
Figure S45. Get High-res Image Peak annotation.

Figure S46. Get High-res Image Peak annotation.
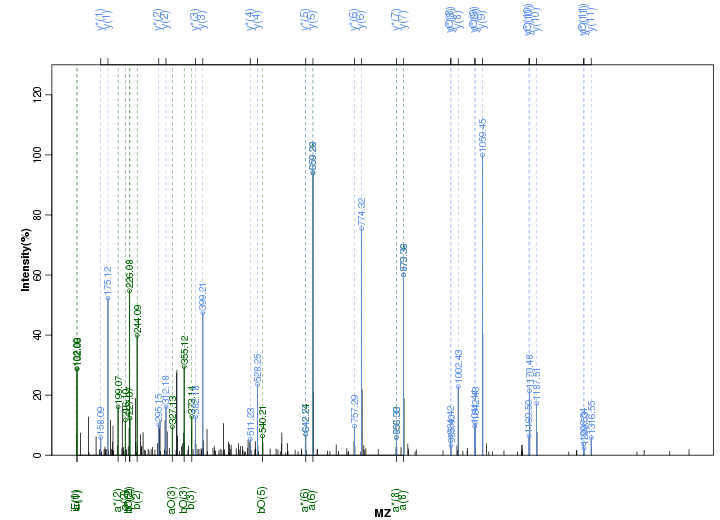
Figure S47. Get High-res Image Peak annotation.
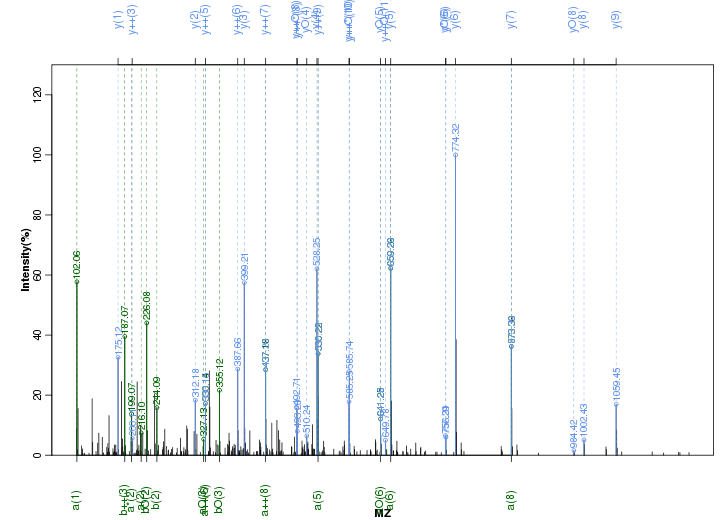
Figure S48. Get High-res Image Peak annotation.
Figure S49. Get High-res Image Peak annotation.
Figure S50. Get High-res Image Peak annotation.
Figure S51. Get High-res Image Peak annotation.
Figure S52. Get High-res Image Peak annotation.
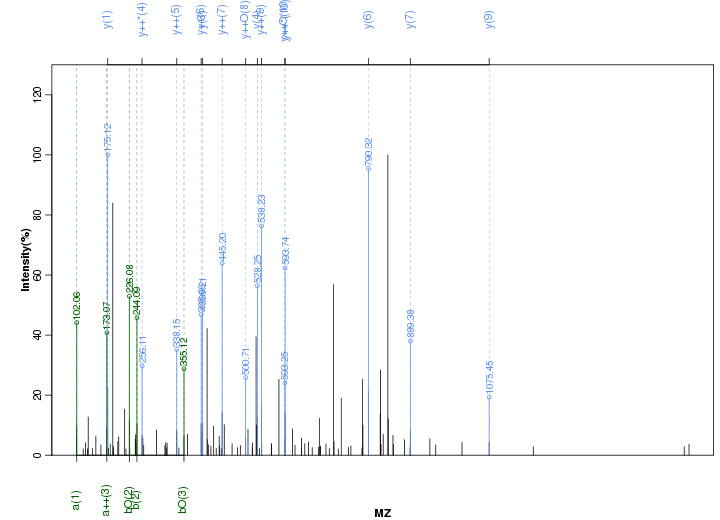
Figure S53. Get High-res Image Peak annotation.
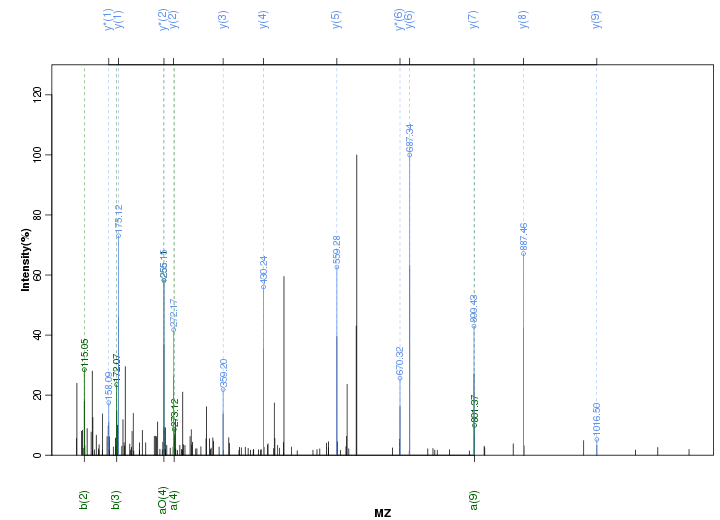
Figure S54. Get High-res Image Peak annotation.

Figure S55. Get High-res Image Peak annotation.

Figure S56. Get High-res Image Peak annotation.
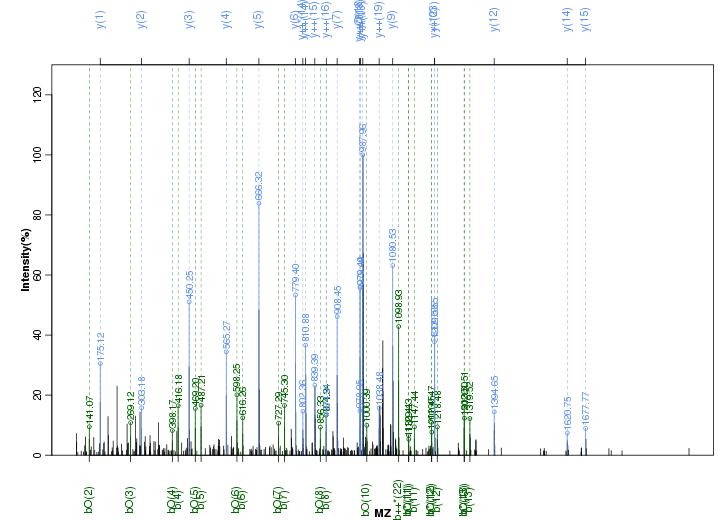
Figure S57. Get High-res Image Peak annotation.
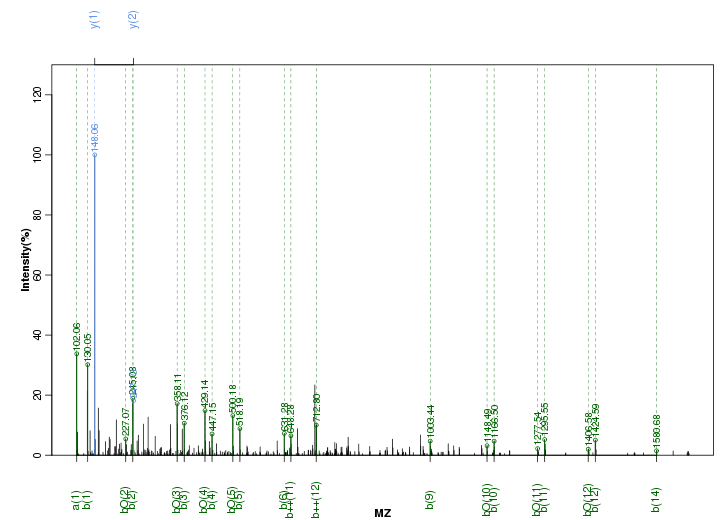
Figure S58. Get High-res Image Peak annotation.
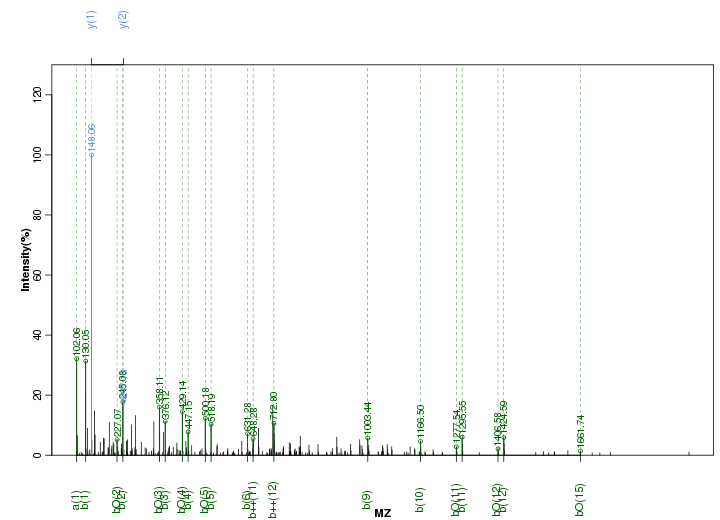
Figure S59. Get High-res Image Peak annotation.
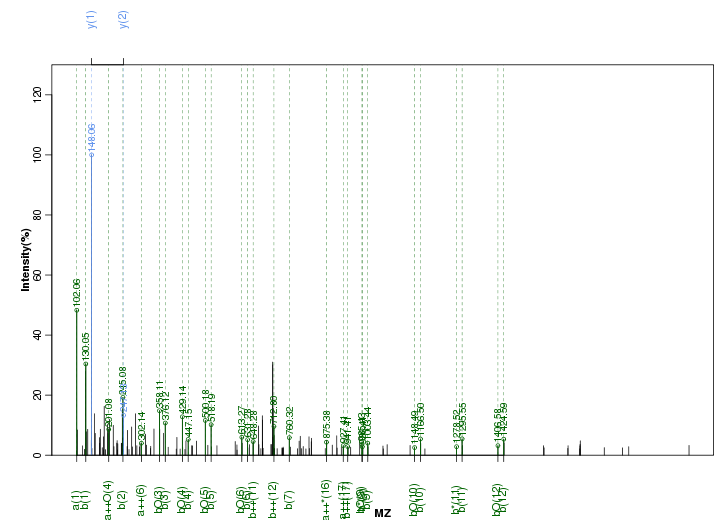
Figure S60. Get High-res Image Peak annotation.

Figure S61. Get High-res Image Peak annotation.
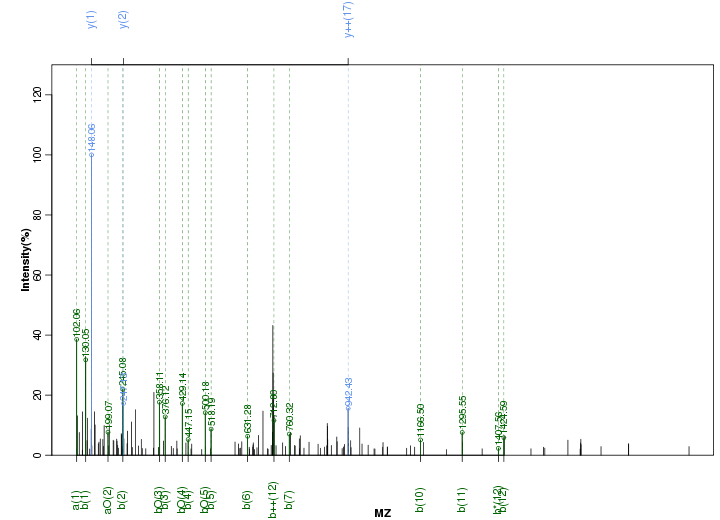
Figure S62. Get High-res Image Peak annotation.

Figure S63. Get High-res Image Peak annotation.

Figure S64. Get High-res Image Peak annotation.
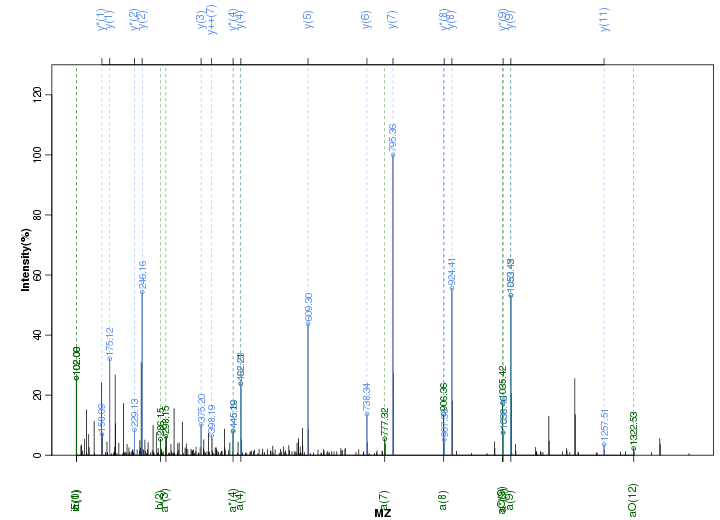
Figure S65. Get High-res Image Peak annotation.
Figure S66. Get High-res Image Peak annotation.
Figure S67. Get High-res Image Peak annotation.
Figure S68. Get High-res Image Peak annotation.
Figure S69. Get High-res Image Peak annotation.

Figure S70. Get High-res Image Peak annotation.
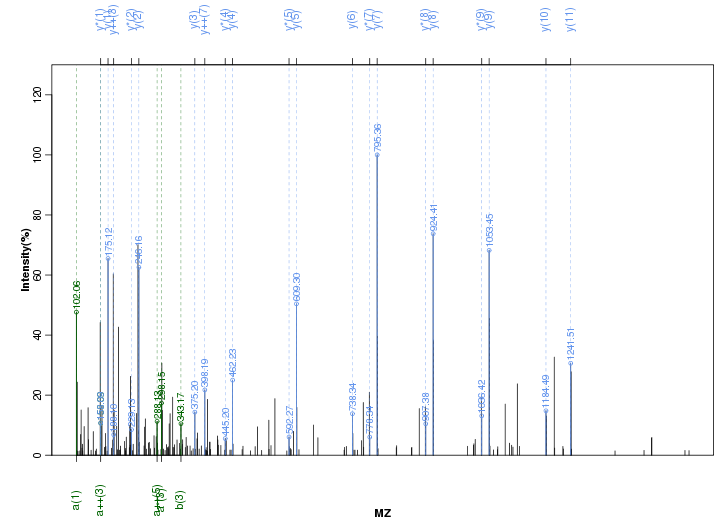
Figure S71. Get High-res Image Peak annotation.
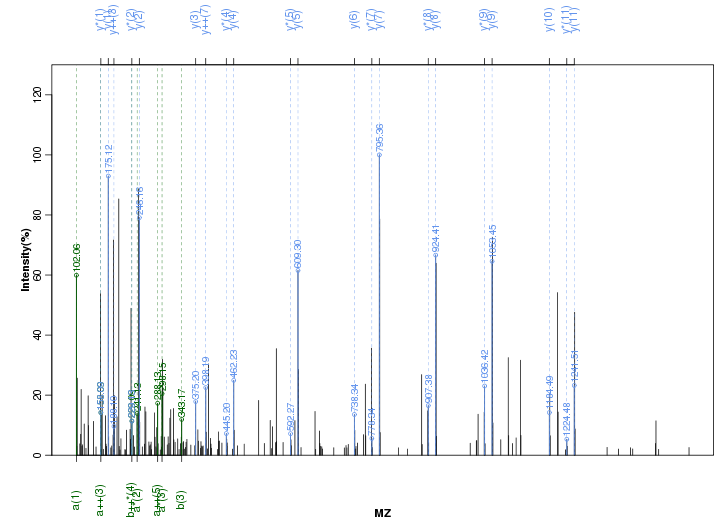
Figure S72. Get High-res Image Peak annotation.

Figure S73. Get High-res Image Peak annotation.
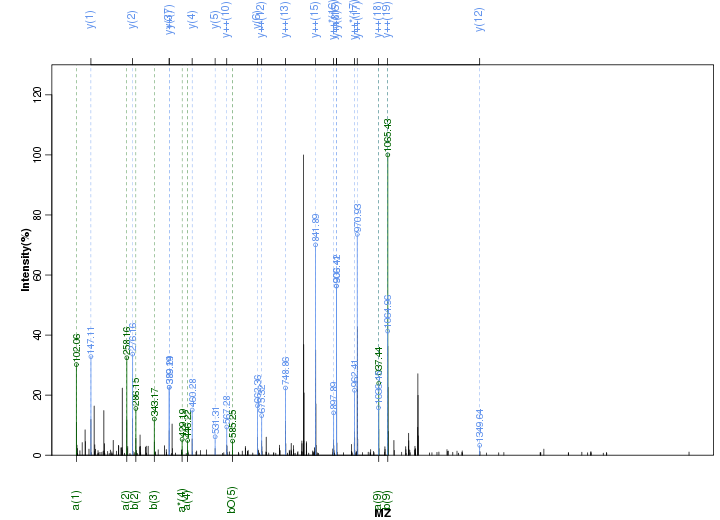
Figure S74. Get High-res Image Peak annotation.
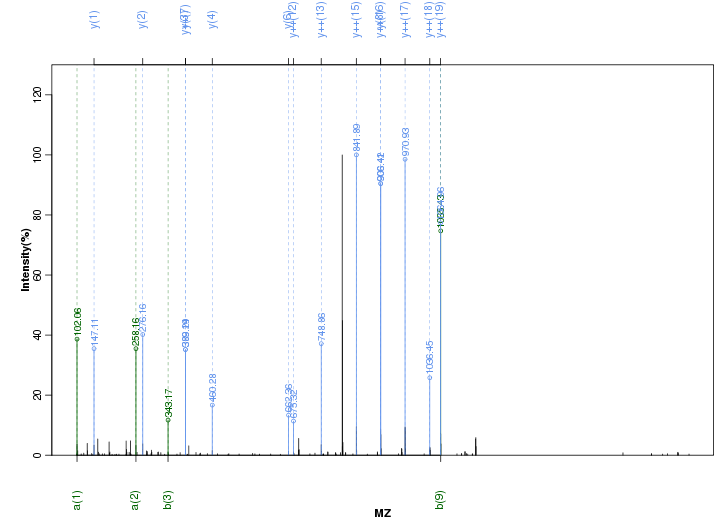
Figure S75. Get High-res Image Peak annotation.
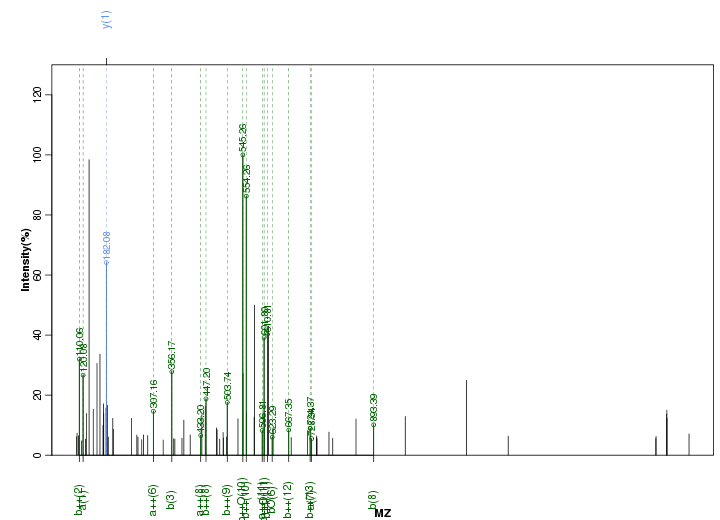
Figure S76. Get High-res Image Peak annotation.
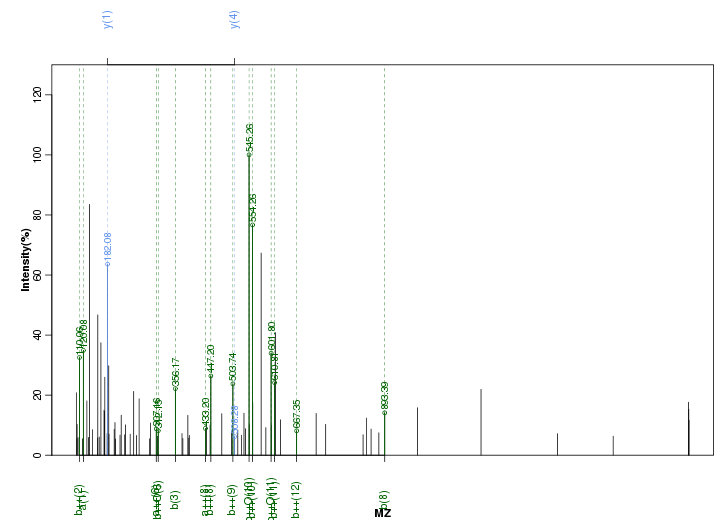
Figure S77. Get High-res Image Peak annotation.
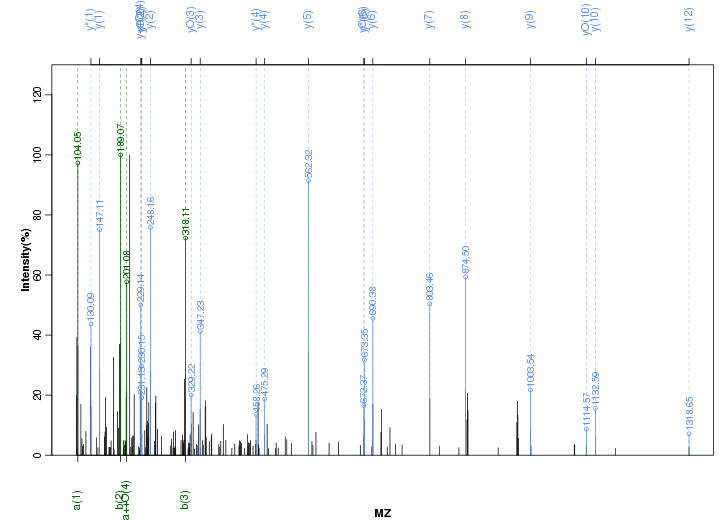
Figure S78. Get High-res Image Peak annotation.

Figure S79. Get High-res Image Peak annotation.

Figure S80. Get High-res Image Peak annotation.
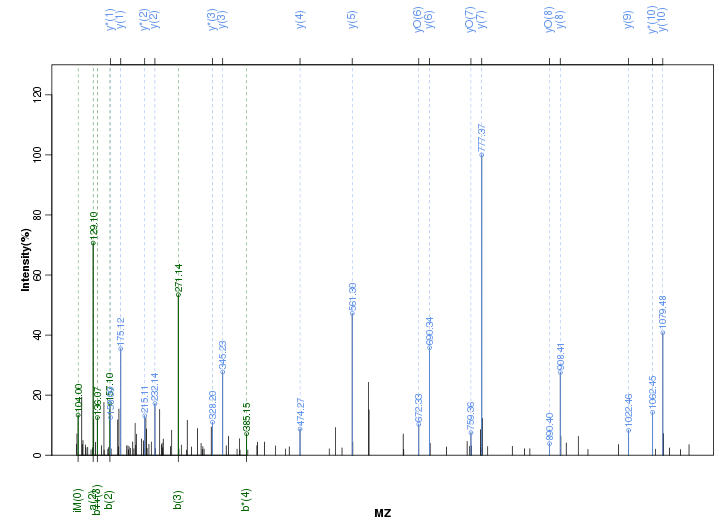
Figure S81. Get High-res Image Peak annotation.

Figure S82. Get High-res Image Peak annotation.
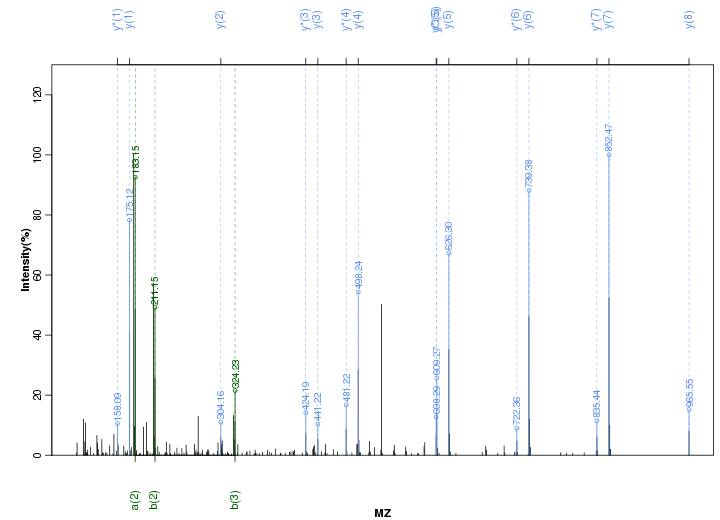
Figure S83. Get High-res Image Peak annotation.
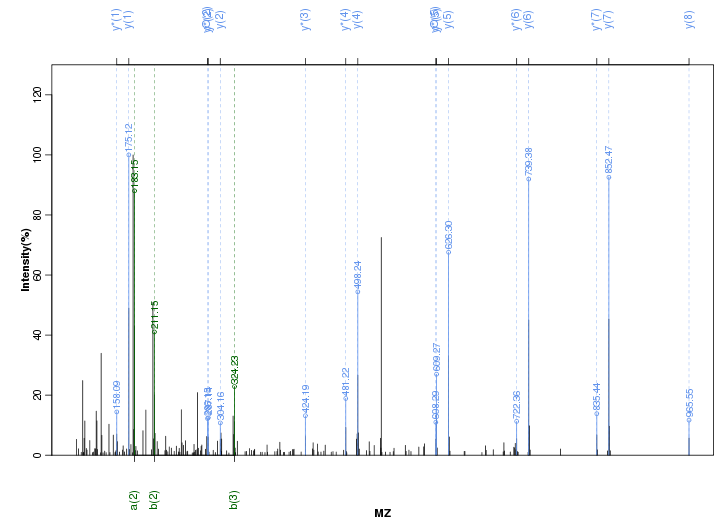
Figure S84. Get High-res Image Peak annotation.
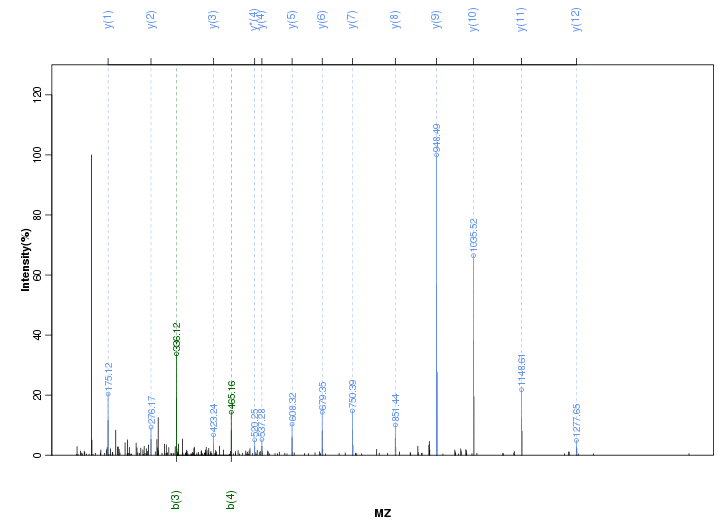
Figure S85. Get High-res Image Peak annotation.
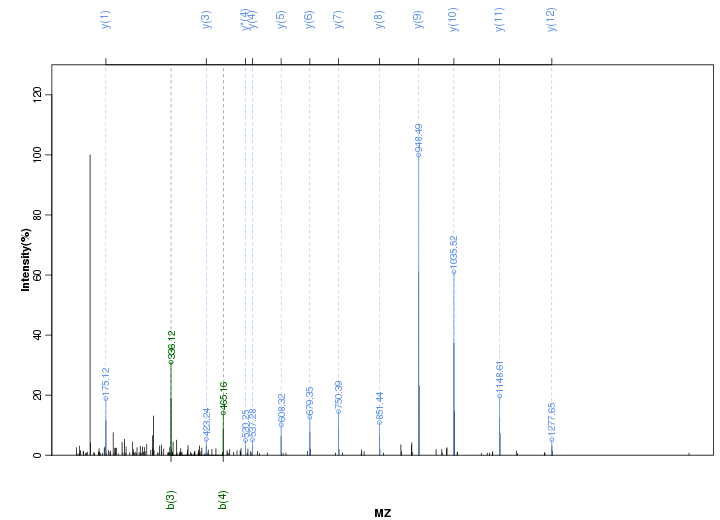
Figure S86. Get High-res Image Peak annotation.
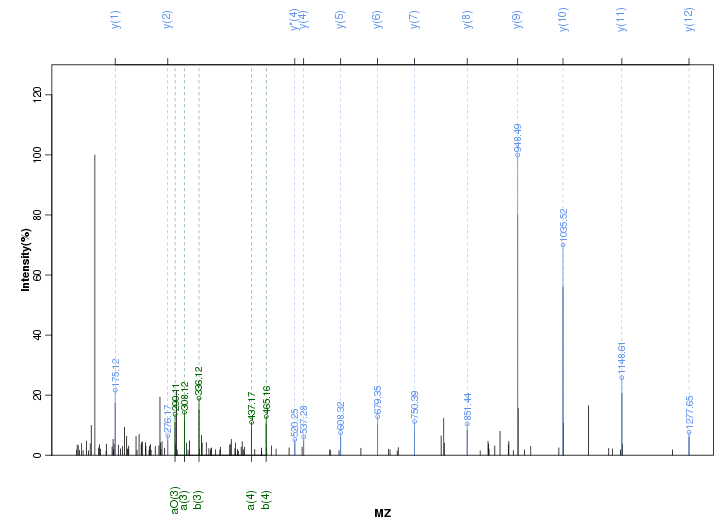
Figure S87. Get High-res Image Peak annotation.
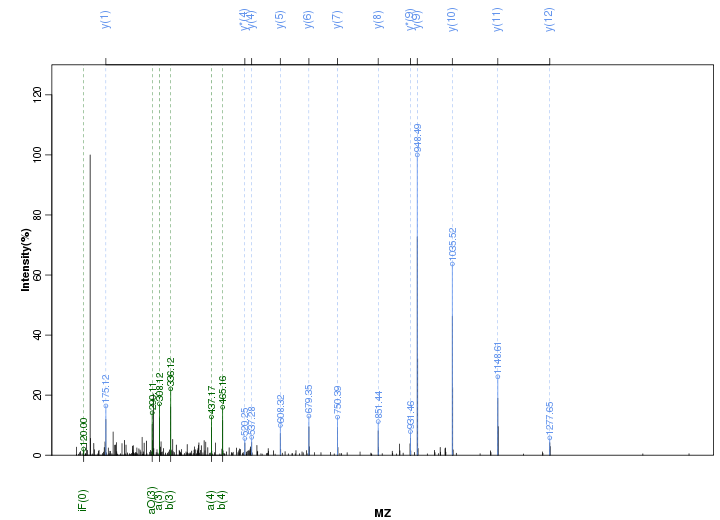
Figure S88. Get High-res Image Peak annotation.
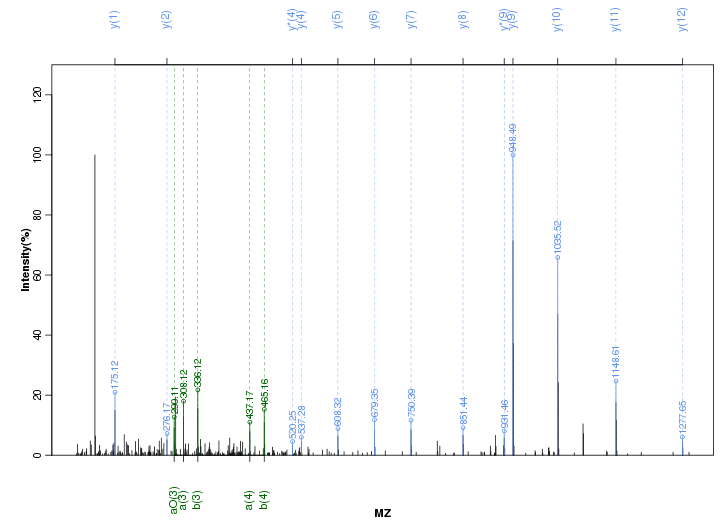
Figure S89. Get High-res Image Peak annotation.

Figure S90. Get High-res Image Peak annotation.
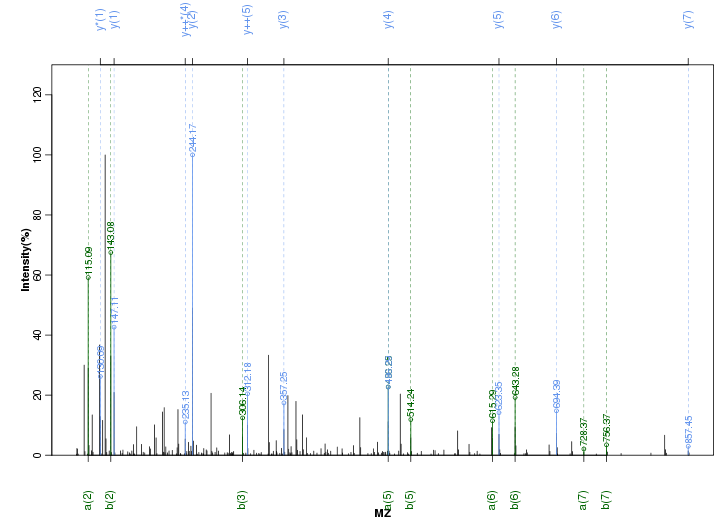
Figure S91. Get High-res Image Peak annotation.
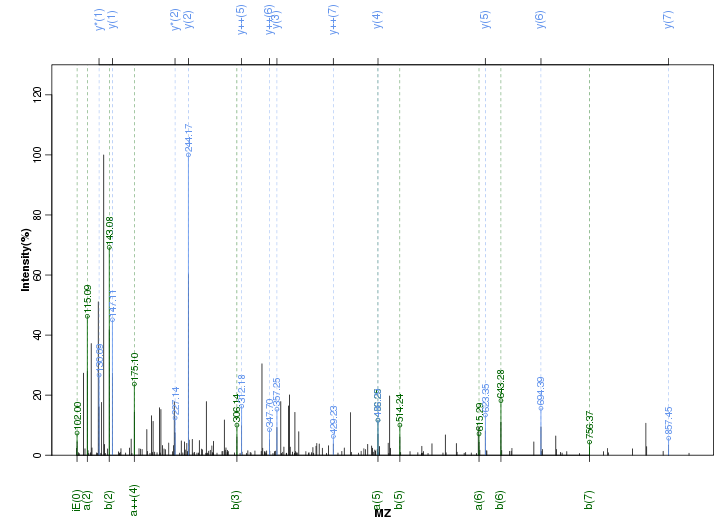
Figure S92. Get High-res Image Peak annotation.
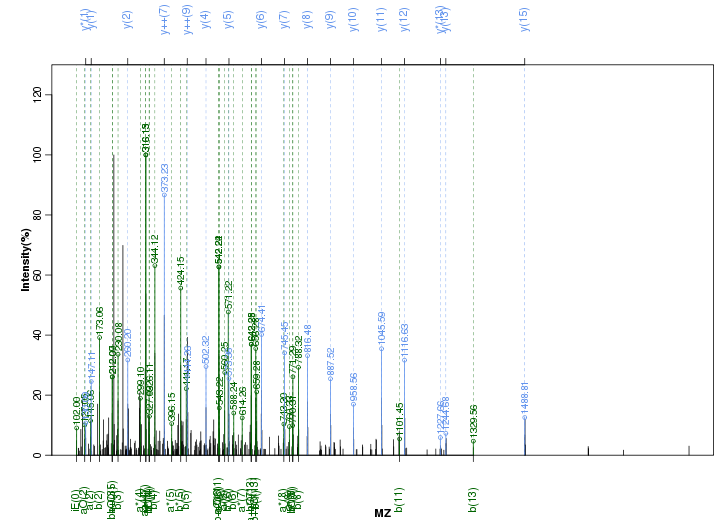
Figure S93. Get High-res Image Peak annotation.
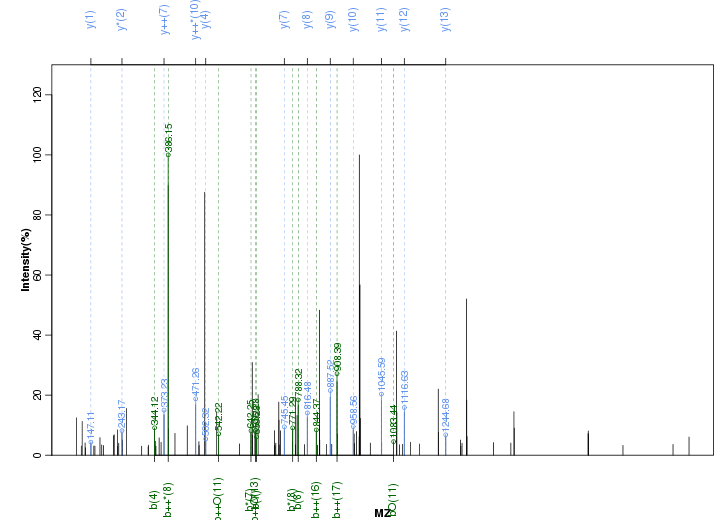
Figure S94. Get High-res Image Peak annotation.
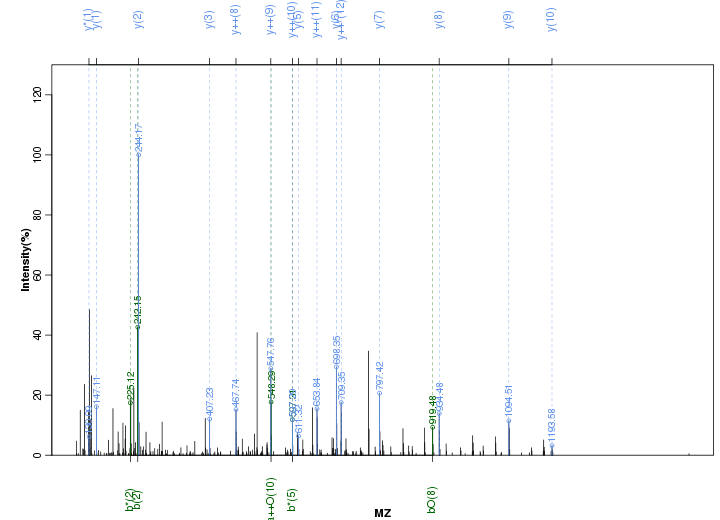
Figure S95. Get High-res Image Peak annotation.
Figure S96. Get High-res Image Peak annotation.

Figure S97. Get High-res Image Peak annotation.
Figure S98. Get High-res Image Peak annotation.
Figure S99. Get High-res Image Peak annotation.
Figure S100. Get High-res Image Peak annotation.
Figure S101. Get High-res Image Peak annotation.
Figure S102. Get High-res Image Peak annotation.
Figure S103. Get High-res Image Peak annotation.
Figure S104. Get High-res Image Peak annotation.
Figure S105. Get High-res Image Peak annotation.
Figure S106. Get High-res Image Peak annotation.

Figure S107. Get High-res Image Peak annotation.

Figure S108. Get High-res Image Peak annotation.

Figure S109. Get High-res Image Peak annotation.
Figure S110. Get High-res Image Peak annotation.
Figure S111. Get High-res Image Peak annotation.
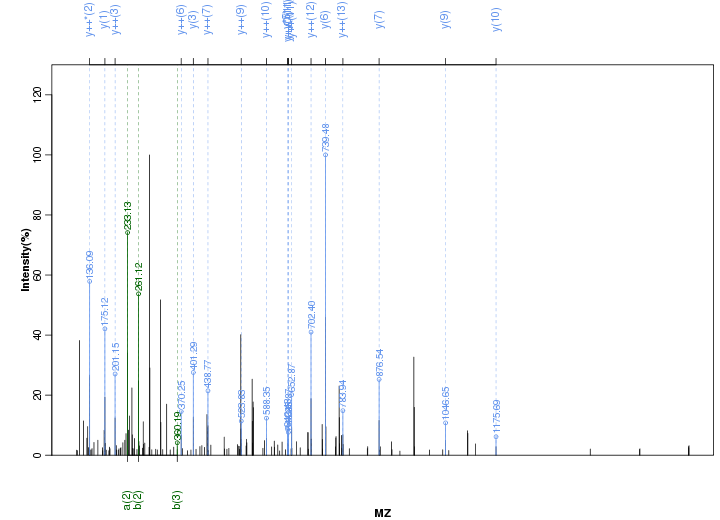
Figure S112. Get High-res Image Peak annotation.
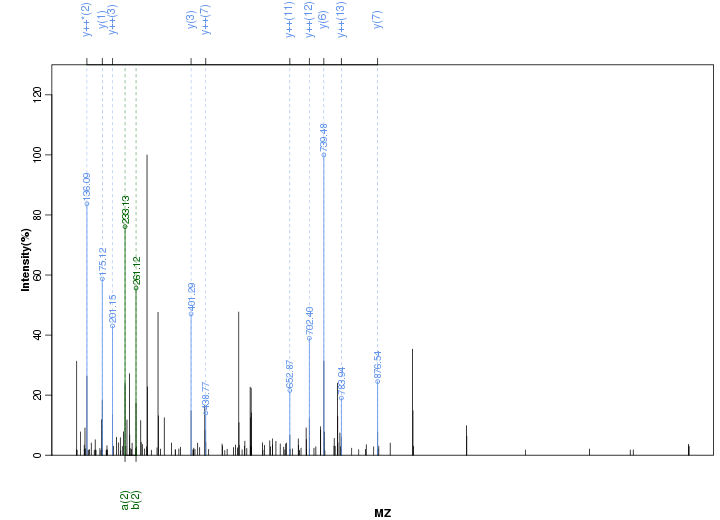
Figure S113. Get High-res Image Peak annotation.
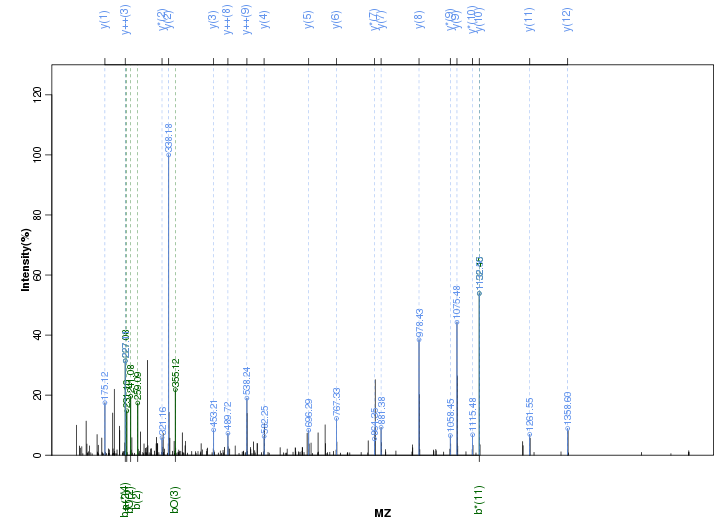
Figure S114. Get High-res Image Peak annotation.

Figure S115. Get High-res Image Peak annotation.

Figure S116. Get High-res Image Peak annotation.

Figure S117. Get High-res Image Peak annotation.
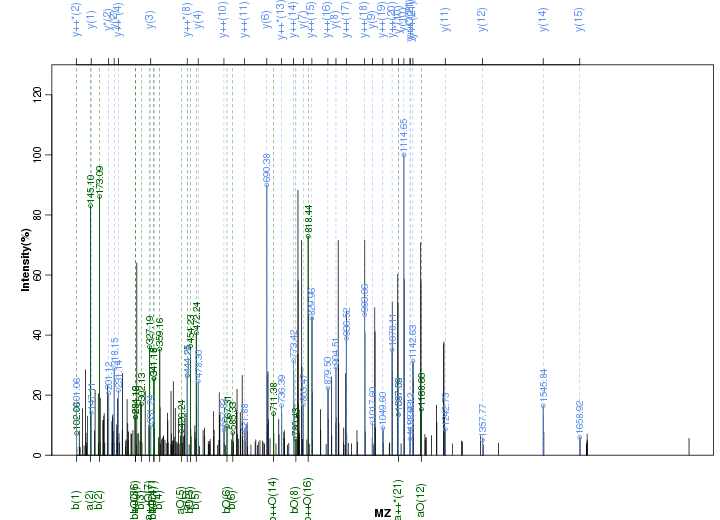
Figure S118. Get High-res Image Peak annotation.
Figure S119. Get High-res Image Peak annotation.
Figure S120. Get High-res Image Peak annotation.
Figure S121. Get High-res Image Peak annotation.
Figure S122. Get High-res Image Peak annotation.
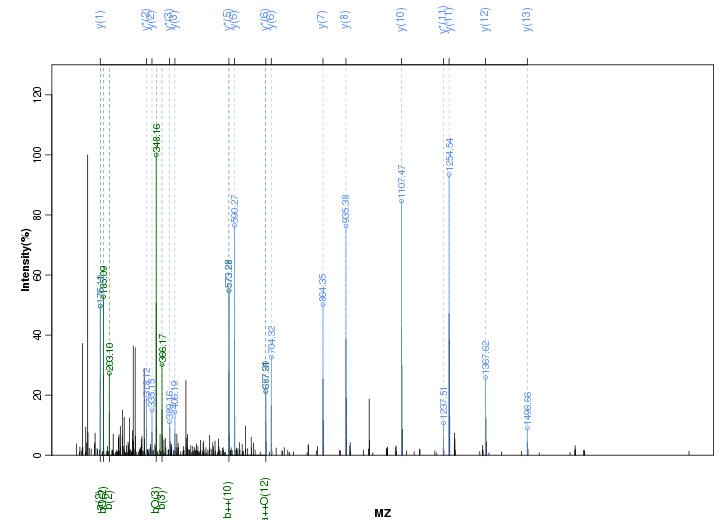
Figure S123. Get High-res Image Peak annotation.